Automatic Diagnosis and Grading of Prostate Cancer with Weakly Supervised Learning on Whole Slide Images
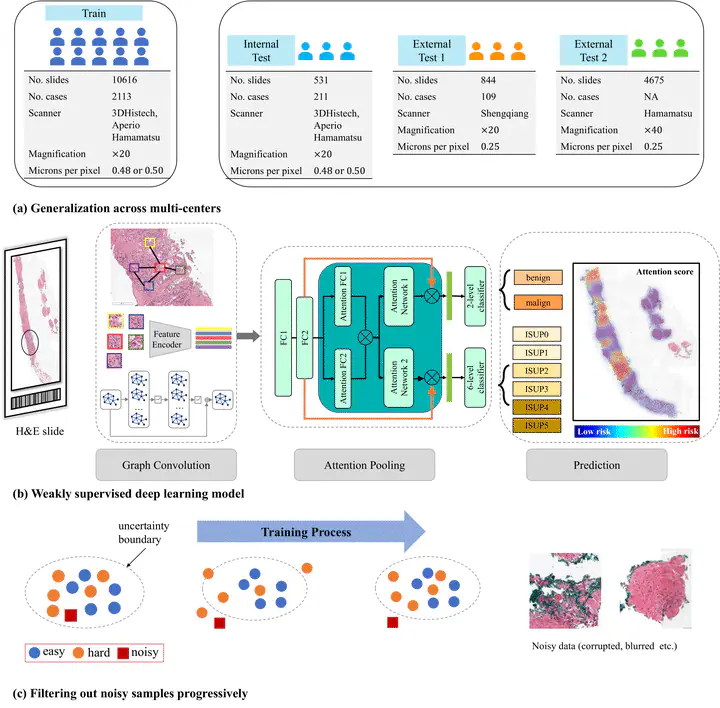 The workflow of automated prostate cancer diagnosis and grading system
The workflow of automated prostate cancer diagnosis and grading systemAbstract
Background. Prostate cancer diagnosis and grading workflow is cumbersome, and the results suffer from substantial inter-observer variability. Recent trials have shown potential in using machine learning to develop automated systems to address this challenge. Most automated deep learning systems for prostate cancer Gleason grading focused on supervised learning requiring demanding fine-grained pixel-level annotations. Methods A weakly supervised deep learning model with slide-level labels is presented in this study for diagnosing and grading prostate cancer with whole slide images (WSI). WSIs are first cropped into small patches and then processed with a deep-learning model to extract patch-level features. A graph convolution network (GCN) aggregates the features for classifications. The noisy labels are progressively filtered out throughout the training process to reduce inter-observer variations in clinical reports. Finally, multi-center independent test cohorts \textcolor{red}{with 6,174 slides are collected to evaluate the prostate cancer diagnosis and grading performance of our model. Results The cancer diagnosis (2-level classification) results on two external test sets (n=4,675, n=844) show an area under the receiver operating characteristic curve (AUC) of 0.985 and 0.986. The results of the Gleason grading (6-level classification) reach 0.931 quadratic weighted kappa on the internal test set (n=531). It generalizes well} on the external test dataset (n=844) with an independent 0.801 quadratic weighted kappa with the reference standard set. The model enables pathological meaningful interpretability by visualizing the most attended lesions, which are highly consistent with expert annotations. Conclusion The proposed model incorporates a graph network in weakly supervised learning with only slide-level reports. A robust learning strategy is also employed to correct the label noise. It is highly accurate $>0.985$ AUC for diagnosis) and also interpretable with intuitive heatmap visualization. It can be unified with a digital pathology pipeline to deliver prostate cancer metrics for a pathology report.