Exploring Low-Rank Property in Multiple Instance Learning for Whole Slide Image Classification
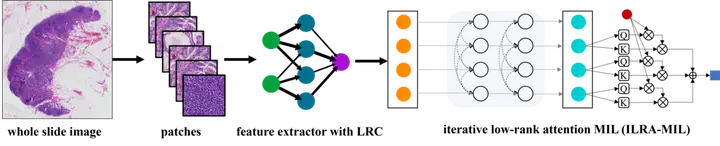 The proposed WSI classification pipeline.
The proposed WSI classification pipeline.Abstract
The classification of gigapixel-sized whole slide images (WSIs) with slide-level labels can be formulated as a multiple-instance-learning (MIL) problem. State-of-the-art models often consist of two decoupled parts local feature embedding with a pre-trained model followed by a global feature aggregation network for classification. We leverage the properties of the apparent similarity in high-resolution WSIs, which essentially exhibit low-rank structures in the data manifold, to develop a novel MIL with a boost in both feature embedding and feature aggregation. We extend the contrastive learning with a pathology-specific Low-Rank Constraint (LRC) for feature embedding to pull together samples (i.e., patches) belonging to the same pathological tissue in the low-rank subspace and simultaneously push apart those from different latent subspaces. At the feature aggregation stage, we introduce an iterative low-rank attention MIL (ILRA-MIL) model to aggregate features with low-rank learnable latent vectors to model global interactions among all instances. We highlight the importance of instance correlation modeling but refrain from directly using the transformer encoder, considering the complexity. ILRA-MIL with LRC pre-trained features achieves strong empirical results across various benchmarks, including (i) 96.49% AUC on the CAMELYON16 for binary metastasis classification, (ii) 97.63% AUC on the TCGA-NSCLC for lung cancer subtyping, and (iii) 0.6562 kappa on the large-scale PANDA dataset for prostate cancer classification.